Even though rootless containers have been a thing for several years now, thanks to the work of Akihiro Suda and others, there is still no community-wide support for using Docker Engine or Podman in the High Performance Computing (HPC) and High Throughput Computing (HTC) communities. Some notable exceptions exist such as the LXPLUS service at CERN, where with some minimal user action (link requires a CERN account) Podman can be used to run OCI containers.
XDG_RUNTIME_DIR=/run/user/$UID upon user login and allocating
subuids and subgids
for each user. In the case of LXPLUS, this is done automatically through a
script
that maps user to subordinate user IDs by shifting the user ID eight bits to the left,
leaving the last 11 bits for the possible subordinate users.
Leveraging containers with Snakemake #
What I actually wanted to do is run a
Snakemake
workflow on my Apple Silicon Mac using containers for each workflow step.
Specifically, I wanted to run a
CMS Higgs boson Open Data example.
In the corresponding
Snakefile,
containers are defined as follows:
rule scram:
container:
"docker://docker.io/cmsopendata/cmssw_5_3_32"
and an example command to run the workflow (from a newly created
snakemake-local-run directory within the cloned repository) is provided as:
snakemake -s ../workflow/snakemake/Snakefile -p --cores 4 --use-singularity \
--configfile ../workflow/snakemake/input.yaml
Looking at snakemake --help, v9.9.0 at the time of writing, I found that
while Snakemake supports
Singularity/Apptainer
natively
and even
Kubernetes
through an
executor plugin,
there is no support for using Docker/Podman.
This is somewhat planned, see the
deployment docs:
other container runtimes will be supported in the future (e.g. podman).
but will probably have to wait until the next Snakemake hackathon.
Apptainer through Lima #
Since spawning a Kubernetes cluster is not something a typical particle physicist will do, I looked into the Apptainer documentation to install Apptainer on Mac:
Apptainer is available via Lima (installable with Homebrew or manually)
I already had Homebrew and Lima installed, and this effectively boils down to
running two commands (one to install brew, the other to install lima)
also listed on the Apptainer webpage.
However, to create the Lima virtual machine (VM) on an Apple Silicon and
analyse
CMS Open Data,
which is using x86 containers, it is
important to pass the
--rosetta flag:
limactl start template://apptainer --cpus 4 --memory 8 --vm-type=vz --rosetta
Should you have forgotten this, you need to delete the VM again:
limactl delete apptainer
Looking at the
apptainer template,
Lima actually provisions a Ubuntu LTS image and runs apt install apptainer
to make apptainer available.
When starting the Lima apptainer instance using limactl shell apptainer,
you will actually only be dumped into a Ubuntu shell.
To directly be able to call apptainer, you can set an alias:
alias apptainer="limactl shell apptainer apptainer"
Now we can e.g. execute the CMS open data container:
apptainer run docker://docker.io/cmsopendata/cmssw_5_3_32
or rather use the image from the CERN GitLab registry (this is what I will use in the following):
apptainer run docker://gitlab-registry.cern.ch/cms-cloud/cmssw-docker/cmssw_5_3_32-slc6_amd64_gcc472
gitlab-registry.cern.ch/cms-cloud/cmssw-docker/al9-cms.
Once the image has been downloaded and converted to the Singularity Image Format (SIF) — which will print a lot of (harmless) warnings — the running of the image will actually fail:
INFO: Inserting Apptainer configuration...
INFO: Adding owner write permission to build path: /tmp/build-temp-1569054307/rootfs
INFO: Creating SIF file...
[===================================================================] 100 % 0s
Setting up CMSSW_5_3_32
perl: warning: Setting locale failed.
perl: warning: Please check that your locale settings:
LANGUAGE = (unset),
LC_ALL = (unset),
LANG = "C.UTF-8"
are supported and installed on your system.
perl: warning: Falling back to the standard locale ("C").
cannot make directory CMSSW_5_3_32 Read-only file system
The reason is that the
entrypoint script
actually tries to create a CMSSW environment.
This fails because the directory where that environment is created is read-only.
Let’s instead get a shell into the container, using shell instead of run:
apptainer shell docker://gitlab-registry.cern.ch/cms-cloud/cmssw-docker/cmssw_5_3_32-slc6_amd64_gcc472
Now we can try to investigate the run error.
I have executed the apptainer command in my home directory:
Apptainer> pwd
/Users/clange
Apptainer> touch test
/bin/touch: setting times of `test': Read-only file system
However, by default, the home directory is mounted as read-only.
The only shared writable directory in the
default configuration
is /tmp/lima.
If we change to that directory, we can actually create and edit files.
Knowing this, we are now able to start (and even run) the container
successfully (I have suppressed all perl locale warnings in the output):
❯ mkdir /tmp/lima/code
❯ apptainer run -u -B /tmp/lima/code:/code docker://gitlab-registry.cern.ch/cms-cloud/cmssw-docker/cmssw_5_3_32-slc6_amd64_gcc472
INFO: Using cached SIF image
Setting up CMSSW_5_3_32
CMSSW should now be available.
This is a standalone image for CMSSW_5_3_32 slc6_amd64_gcc472.
[10:46:57] clange@lima-apptainer /code/CMSSW_5_3_32/src $ cmsRun --help
/cvmfs/cms.cern.ch/slc6_amd64_gcc472/cms/cmssw/CMSSW_5_3_32/bin/slc6_amd64_gcc472/cmsRun [options] [--parameter-set] config_file
Allowed options:
-h [ --help ] produce help message
-p [ --parameter-set ] arg configuration file
-j [ --jobreport ] arg file name to use for a job report file: default
extension is .xml
-e [ --enablejobreport ] enable job report files (if any) specified in
configuration file
-m [ --mode ] arg Job Mode for MessageLogger defaults - default mode
is grid
-t [ --multithreadML ] MessageLogger handles multiple threads - default
is single-thread
--strict strict parsing
Getting Snakemake to work #
It is not very practical to channel all interactions with the container through
the /tmp/lima directory.
Let’s edit the configuration to make the home directory writable.
For this, we first need to stop the VM:
limactl stop apptainer
limactl edit apptainer
The
Lima FAQ on file system sharing
list what to add to the config (the mounts section is close to the end of the
configuration). The - location: "~" is already there, we only need to add the
writable: true line:
mounts:
- location: "~"
writable: true
After saving the file and exiting the editor, Lima will ask if the instance
should be started again, answer with “yes”.
If we now mount the home directory in the apptainer shell command, we can
write to any place in the home directory:
❯ apptainer shell -B $HOME:$HOME docker://gitlab-registry.cern.ch/cms-cloud/cmssw-docker/cmssw_5_3_32-slc6_amd64_gcc472
INFO: Using cached SIF image
Apptainer> pwd
/Users/clange
Apptainer> touch test
Let’s now clone the above-mentioned
CMS Higgs boson Open Data example
and replace the image name by the gitlab-registry.cern.ch one:
git clone https://github.com/reanahub/reana-demo-cms-h4l.git
cd reana-demo-cms-h4l
sed -i '' 's|docker://docker.io/cmsopendata/cmssw_5_3_32|docker://gitlab-registry.cern.ch/cms-cloud/cmssw-docker/cmssw_5_3_32-slc6_amd64_gcc472|g' workflow/Snakefile
We also need to adjust the location of the cmsset_default.sh script
since it is different in that later image,
set an additional environment variable,
and ignore unbound environment variables.
This can be done with the following perl command:
perl -0777 -pe 's/ "source \/opt\/cms\/cmsset_default\.sh "/ "export CMS_PATH=\/cvmfs\/cms.cern.ch "\n "&& set +u; source \/cvmfs\/cms.cern.ch\/cmsset_default.sh; set -u "/g' -i '' workflow/Snakefile
such that the final diff (in 4 places) should be as follows:
container:
- "docker://docker.io/cmsopendata/cmssw_5_3_32"
+ "docker://gitlab-registry.cern.ch/cms-cloud/cmssw-docker/cmssw_5_3_32-slc6_amd64_gcc472"
shell:
- "source /opt/cms/cmsset_default.sh "
+ "export CMS_PATH=/cvmfs/cms.cern.ch "
+ "&& set +u; source /cvmfs/cms.cern.ch/cmsset_default.sh; set -u "
Next, follow the instructions in the
Snakefile:
mkdir snakemake-local-run
cd snakemake-local-run
python3 -m venv .venv
source .venv/bin/activate
pip install snakemake
cp -a ../code ../data .
The snakemake command is actually not quite right, it needs to be:
snakemake -s ../workflow/Snakefile -p --cores 4 --use-singularity \
--configfile ../workflow/input.yaml
and since Snakemake version 8, this should actually be changed to:
snakemake -s ../workflow/Snakefile -p --cores 4 --software-deployment-method apptainer \
--configfile ../workflow/input.yaml
Still, this fails:
Assuming unrestricted shared filesystem usage.
host: mpc3044.clange.ch
Building DAG of jobs...
WorkflowError:
The apptainer or singularity command has to be available in order to use apptainer/singularity integration.
Since Snakemake uses the Python
shutil.which function
to determine the availability of the singularity command, even using
alias singularity=apptainer does not work.
We actually need to have an executable file with that name available.
Let’s create one as ~/bin/singularity with the following content:
#!/bin/zsh
limactl shell apptainer apptainer "$@"
and make sure that it gets picked up.
chmod +x ~/bin/singularity
export PATH=~/bin:${PATH}
Now singularity --version should return the apptainer version number.
And this is actually all we need to execute the workflow successfully.
Depending on the network connection, this can now take around 15 minutes.
Once the workflow has completed, you should see the following files in your
snakemake-local-run/results directory:
total 272
-rw-r--r-- 1 clange staff 54181 Aug 24 12:52 DoubleMuParked2012C_10000_Higgs.root
-rw-r--r-- 1 clange staff 59770 Aug 24 12:35 Higgs4L1file.root
-rw-r--r-- 1 clange staff 18141 Aug 24 12:52 mass4l_combine_userlvl3.pdf
-rw-r--r-- 1 clange staff 0 Aug 24 12:33 scramdone.txt
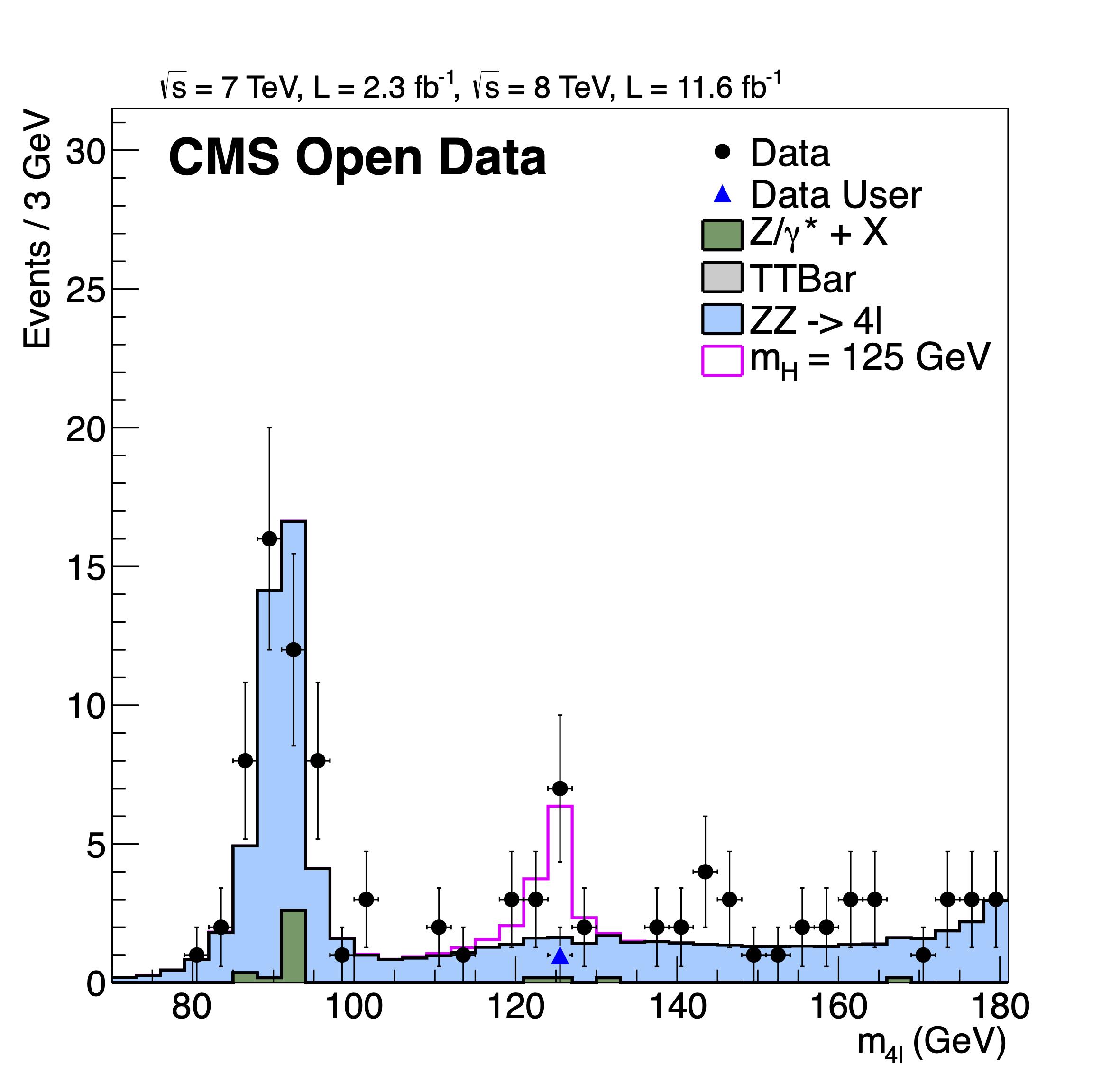
and the resulting mass4l_combine_userlvl3.pdf (here converted into PNG
format) should look as follows: